Why cyclic peptides?
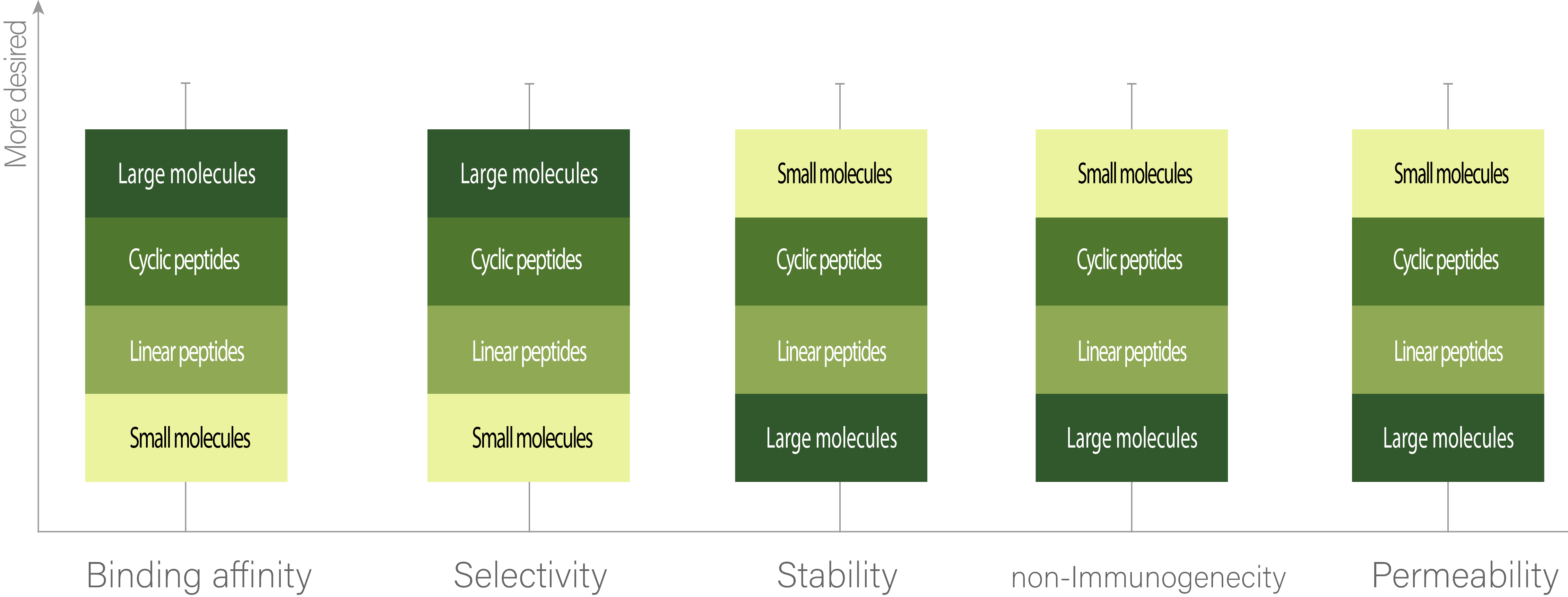
Cyclic peptides are an emerging category of therapeutics. There are several attributes that make cyclic peptides a desirable therapeutic agent. In Fig. 1, some of these drug-favorable properties are compared among different categories of therapeutics. As it can be seen, cyclic peptides can have high binding affinity and selectivity without losing stability or increasing immunogenic responses. This mostly is related to their size, which lies in between small molecules (<500 Da) and large molecules (>5000 Da), and allows them to retain both the rigidity of small molecules for stability and less immunogenicity and wide surface area of large molecules for better binding affinity. In terms of permeability, even though many peptides (cyclic or linear) can not pass the cell membrane, there still are some cyclic peptides, like Cyclosporine A, that have a high permeability. Also, generally, it’s been shown that cyclic peptides are more permeable than their linear counterparts.
- Memebrane permeability prediction
Membrane permeability comes to focus when the target is an intracellular protein. Notably, the majority of undrugged protein-protein interaction (PPI) sites are inside the cell, and cyclic peptides are one of the most promising candidates for targeting them. Therefore, being able to find and design permeable cyclic peptides would be a critical step in drug discovery.
In my research, I want to fill in these gaps, respectively, by sampling a diverse set of peptides that are well representative of the whole space and then making a predictive model using machine learning approaches that can easily be used for every cyclic peptide. And this model will contribute to the machine learning field as well because of some unique properties of this problem, like circularity of the sequence and sparcity of the data.
So far, I have made a diverse set of peptides by statistical modeling, and in the next step, after getting experimental results, we will going to train the model.
-
Structure ensemble prediction In this project, we are generating an ensemble of structures for each sequence of cyclic peptide, using Rosetta GenKIC app (aka. simple_cycpep), to use it as the training set for prediction model with machine learning algorithms. The data is alreasy generated and clustered for 50,000 sequences, and in the next step we are going to train the model.
-
Permeability assay This information on this part in confidential and I can not share it before publication. This is going to be a new experimental on permeability of peptides, either cyclic or linear.